Researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences, in collaboration with their partners, have developed a novel method for the comprehensive evaluation of probiotic products using a Raman Flow Cytometry (RFC) platform.
Published in iMetaOmics, this new method allows for rapid, label-free analysis of identity, viability, and metabolic activity at both the single-cell and strain levels in complex probiotic products, creating a "microbial ID card" for each formulation.
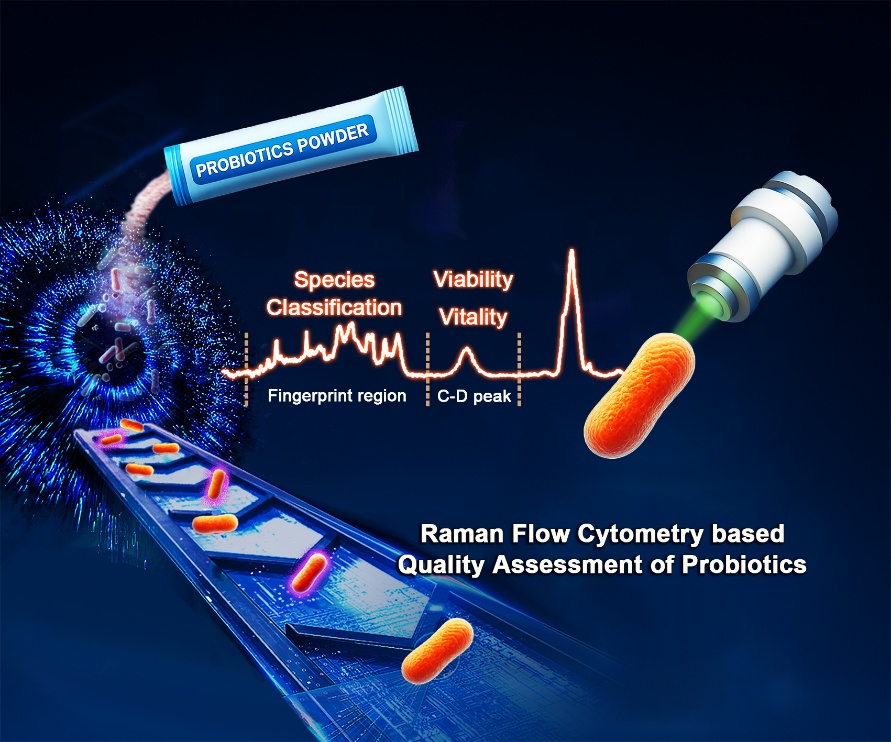
A new Raman Flow Cytometry–based technology for probiotic quality assessment. (Image by LIU Yang)
Ensuring the accuracy and functionality of probiotic formulations is essential for their efficacy and safety. However, traditional methods such as culture-based enumeration and molecular assays can be time-consuming, labor-intensive, and limited in resolution.
To address these challenges, the researchers created the RFC-based method, which integrates high-throughput Raman spectroscopy with microfluidics and machine learning. This new system captures spontaneous Raman spectra from individual cells at a speed of one cell per second, achieving more than ten times higher throughput than previous Raman-based methods. A curated spectral reference database, combined with a linear discriminant analysis model, allows for precise classification of probiotic strains, including the detection of those present at just 0.1% abundance.
In addition to strain identification, the researchers evaluate both cell viability and metabolic activity using deuterium-labeled water (D2O) as a metabolic tracer. The incorporation of D2O into cellular biomolecules produces characteristic Raman shifts, enabling non-invasive quantification of each cell's physiological state without the need for fluorescent labeling.
The RFC workflow was validated across both single-strain and multi-strain commercial probiotic products, achieving results comparable to traditional plate-counting and sequencing methods. Compared to these conventional tools, RFC significantly reduces assay time and complexity while providing deeper insights into strain-level vitality and intercellular heterogeneity.
"The RFC platform demonstrates how in-depth single-cell analysis can be transformed into practical tools for industrial microbiology," said co-first author Prof. ZHANG Jia from QIBEBT. "This method bridges fundamental microbial phenotyping with real-world quality assurance, offering both scientific rigor and operational value."