Chinese researchers have developed a new platform that cuts the time to diagnose bloodstream bacterial infections and identify effective antibiotics from days to just 12 hours, according to a recent study published in Sensors and Actuators B: Chemical.
A microfluidic chip-based platform enables rapid detection of pathogens. (Image by LIU Yang)
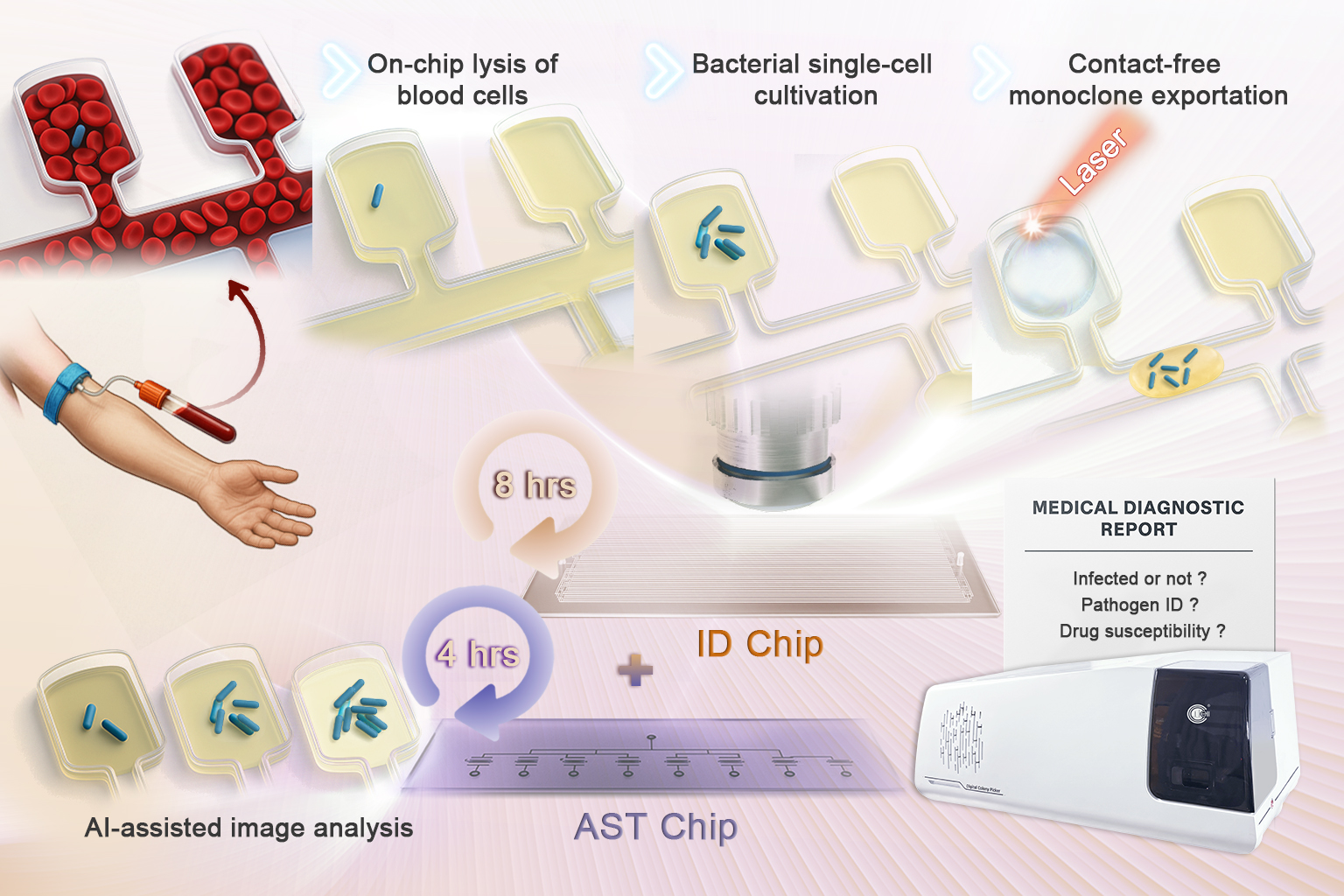
A research team from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences, collaborating with other researchers, created the system to address a critical gap in global healthcare: bloodstream infections (BSIs) are a leading cause of sepsis and death, yet traditional diagnostic methods—relying on pathogen culturing from blood—often take 48 to 72 hours, delaying life-saving treatment.
The new platform integrates two microfluidic Static Droplet Array (SDA) chips: an identification (ID) chip and an antimicrobial susceptibility testing (AST) chip. This combination streamlines the entire workflow from pathogen detection and identification to determining which antibiotics will work.
The ID chip isolates individual bacteria into more than 50,000 nanoliter-scale chambers. Using a metabolism-responsive dye, the system cultures the bacteria and enables fluorescence-based detection within three to five hours, with a sensitivity to detect as few as 10 colony-forming units (CFU) per milliliter.
Next, laser-induced microbubbles release microcolonies, which are then identified via matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry.
The AST chip then exposes the identified bacteria to multiple antibiotics simultaneously in microchambers. An AI-assisted imaging system tracks bacterial growth and calculates minimum inhibitory concentrations (MICs) for each antibiotic in under five hours.
While the platform has not yet been tested on direct clinical blood samples, it was validated using two types of specimens: blood spiked with four common BSI-causing bacteria (E. coli, S. aureus, A. baumannii, and K. pneumoniae), and a cultured E. coli isolate from a clinical blood sample provided by the Affiliated Hospital of Qingdao University. In the latter test, MIC results matched those from standard laboratory methods.
"Our system integrates key diagnostic steps into one platform and offers rapid, quantitative results using only small sample volumes," said Dr. DIAO Zhidian, the lead author of the study. "While clinical blood samples were not tested directly, our use of both spiked samples and cultured isolates shows promising technical readiness."