The work was published in Sensors and Actuators B: Chemical on Jan. 12.
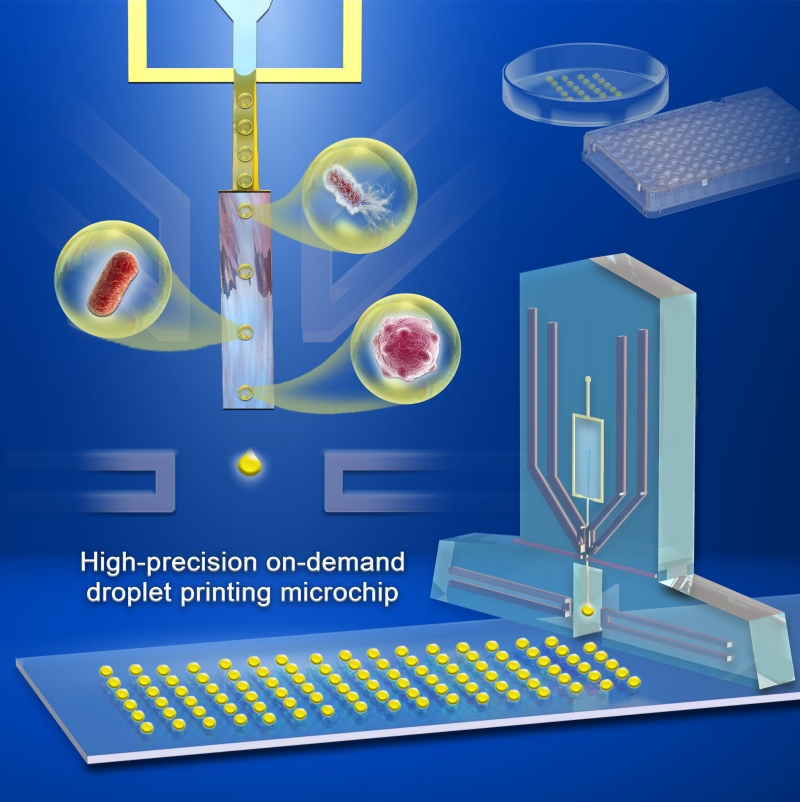
Droplet printing platform for single-cell phenotype screening
(Image by LIU Yang)
Agar plate and microplate screening are the two most widely used traditional screening methods. Agar plate assays are easy to operate but lack precise quantification and are therefore mainly used for initial strain screening. Microplate assays provide accurate quantification and are suitable for evaluating mutant performance, but have lower throughput.
Droplet microfluidics-based tools hold great promise for ultra-high throughput screening. However, current sorting strategies often only enrich droplets with a particular phenotype in a single channel, making it challenging to precisely and efficiently separate each 'target droplet' to specific locations within macroscopic media (e.g., culture dishes or 96-well plates) for subsequent single-cell culture and sequencing experiments.
The droplet printing platform developed in this study consists of four main modules: a pump-driven microfluidic chip module, an automatic optical signal recognition module, a high-pressure control module, and an automated mobile platform module.
DIAO Zhidian, a PhD student and co-first author of the study, explained that the chip contains embedded optical fibers that can introduce red light-emitting diodes. When the sorted target droplets pass through the detection window, the photodiodes connected to the optical fibers can identify them and generate corresponding signals. These signals are then used to print the droplets in specific areas based on electrohydrodynamic principles.
"This chip platform has several unique features," said GE Anle,co-first author of the study. "Not only can this platform generate droplets online, but it can also accurately print incubated or sorted droplets accurately on demand, converting 'target droplet groups' into 'individual target droplets,' thereby serving downstream phenotype studies at the single cell level."
"This platform incorporates an optical detection module based on optical fiber transmission," said co-corresponding author Prof. MA Bo. "This module not only improves the accuracy of droplet printing, but also avoids complex optical path systems. This concise and integrated system is compatible with a wider range of downstream experimental scenarios, such as mass spectrometry, cell culture dishes, and 96-well plates."
Next, the research team plans to integrate this high-precision on-demand single-droplet printing platform with the previously developed Raman-activated cell sorting instruments, such as FlowRACS and RACS-Seq/Culture, so as to achieve fully automated, high-precision, high-throughput functional single-cell identification, separation, culture, and export for complex biological systems such as microbiomes and large-scale mutant libraries.
"The integration of our platform with existing single-cell Raman sorting instruments such as FlowRACS will open up new possibilities for large-scale single-cell functional studies, providing a comprehensive solution for biobanking, synthetic biology, precision medicine, deep-sea/deep-earth/deep-space exploration, and biosecurity," said Prof. XU Jian, head of the Single-Cell Center of QIBEBT.