Cells are a little easier to understand, thanks to improved technology developed by a team of researchers based in China. Using a method called Raman-Activated Cell Ejectionand Sequencing (RACE-Seq), the scientists were able to greatly improve the success of identifying and sequencing individual cells from our environments to understand the cells’ functions.
They published their results recently in Analytical Chemistry.
The procedure of Raman-Activated Cell Ejectionand Sequencing (RACE-Seq)
(Image by SU Xiaolu)
"RACE-Seq is a useful technology to identify, isolate and sequence single-cells with particular function from environment," said SU Xiaolu, paper author and a researcher with the Single-Cell Center in Qingdao Institute of Bioenergy and Bioprocess Technology(QIBEBT), Chinese Academy of Sciences(CAS). "However, the success rate and quality of RACE-Seq has been quite low for environmental samples."
Single-cell Raman sorting and sequencing tools such as RACE-Seq help researchers understand the mechanistic links between the function and genetic components of individual cells in nature. For genetically varied environments, such as soil found in the environment, RACE-Seq allows the cells to be sorted based on their function roles– without damaging them– and have their genome sequenced to establish those mechanic links between genetics and function.
During RACE-Seq, cells are loaded onto a microchip surface and air-dried immediately before being irradiated with a laser. Cells identified with the function being sorted for are identified and ejected into a receiving well on another chip. The cells are then broken apart and their genetic material processed to determine the underlying mechanical link to the particular function of interest.
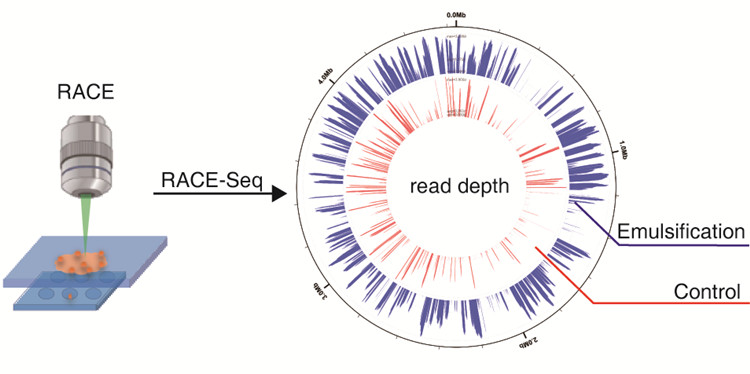
Previously, this method can produce about 20% coverage of genome for individual E. coli cells. Now, the researchers have improved the method so the coverage reaches around 50%.
SU and his team found that adjusting the laser’s energy input helped protect the cells to have better output. They also found that changing the duration, temperature or other aspects of the other steps did not help improve the output.
To tackle this problem, the researchers added oilbefore amplifying the DNA of sorted cells. This simple yet elegant operation reduces such harmful effects of Raman irradiation and increases greatly the completeness of the genomes. Moreover, for soil microbiota, which are perhaps the most complex microbial communities in nature, this new protocol dramatically improves experimental success rate of RACE-Seq.
"Our findings provide a practical solution for enhancing RACE-Seq performance, andthus should make this technique more accessible to the many laboratories interested in single-cell sequencing around the globe," saidXU Jian, director of the Single-Cell Center.
The researchers also noted that RACE-Seq is still unable to produce good-quality genomes at precisely one-cell resolution from environmental samples. They are introducing novel technologies that can achieve the ultimate goal of functionally sorting and sequencing every microbial cell from natural environments.
This work was supported by Chinese Academy of Sciences, Qingdao National Laboratory for Marine Science and Technology, and the National Natural Science Foundation of China.