Efficient biological degradation of cellulosic biomass has been recognized as one major bottleneck in production of cellulosic liquid fuels or biogas, and as the one key step in the carbon cycle of biosphere. In a new work published online in Nature Communications on 24 April, 2015, a research team from Single-Cell Center of Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CAS-QIBEBT) discovered a new mechanism in microbial degradation of lignocellulose, by reporting that cellulosome stoichiometry is regulated by selective RNA processing and stabilization (SRPS).
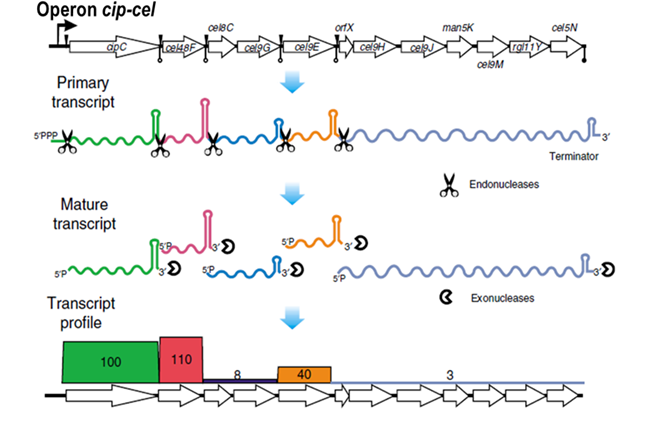
Figure 1 A model for regulation of the stoichiometry of cellulosomal components in vivo.
Cellulosome is the most efficient machinery for cellulose degradation found in nature, and is a high-molecular-mass protein complex assembled from extracellular enzymes. To achieve the proper function, it is crucial to maintain appropriate stoichiometry among these cellulosomal subunits. However, how do cells encode, recognize and control the information?
By mapping the genome-wide transcriptional start sites (TSs) and post-transcriptional processed sites (PSs) of transcripts via a differential mRNA-sequencing approach (dRNA-Seq), Dr XU Chenggang, graduate student HUANG Ranran and their colleagues from Functional Genomics Group of Single-Cell Center, CAS-QIBEBT revealed that in the cellulolytic model organismClostridium cellulolyticum, SRPS mechanism precisely regulates cellulosome stoichiometry at the post-transcriptional level.
The team found that the cip-cel cluster which encodes 12 cellulsomal subunits is transcribed as a single transcriptional unit, namely an “operon”. However, these genes exhibit highly skewed transcript abundance, with a ratio of 100:110:9:8:38:5:4:2:3:2:3:5. This ratio of transcript abundance is correlated with the relative abundance at the protein level. The team revealed that at least five PSs located in the intergenic regions of the cip-cel operon are specifically recognized by endonucleases, resulting in cleavage of the primary transcript into at least six secondary transcripts. Stability of these secondary transcripts varied widely due to their distinct terminal stem-loop structures, which quantitatively determine the observed stoichiometry (Figure 1). Intriguingly, orthologous stem-loops were found in the intergenic regions of cip-cel-like operons from other Clostridia. Therefore, the PSs and the stem-loops precisely regulate structure and abundance of the subunit-encoding transcripts processed from a primary polycistronic RNA, quantitatively specifying a cellulosome “recipe”, and drive evolution of bacterial cellulose degradation.
This simple and ingenious mechanism that controls stoichiometry of protein complex not only ensures simultaneous precise control of ingredients of cellulosome “recipe” following a certain ratio, but also allows reliable inheritance of the “recipe”. Moreover, this discovery introduces a new approach to construct “Super Cellulosome” and “Super Cellulolytic Bacteria” and might enable applications in design and engineering of synthetic protein-machineries.
This work was a collaborative work byFunctional Genomics Groupled by Dr XU Jian and Metabolomics Group led by CUI Qiu, both from CAS-QIBEBT. It was funded by grants from the Ministry of Science and Technology of China and the National Natural Science Foundation of China.