A new platform established by researchers at the Single-Cell Center, Qingdao Institute of Bioenergy and Bioprocess Technology of the Chinese Academy of Sciences (QIBEBT/CAS) improves accuracy, throughput, and stability in profiling dynamic metabolic features of cells – the basic building blocks of all life forms on Earth.
Their study, published in Advanced Science on Mar. 4, describes how this technology allows scientists to take metabolism-based snapshots of cell populations, including plants (microalgae), yeast, bacteria like E. coli and human cancers.
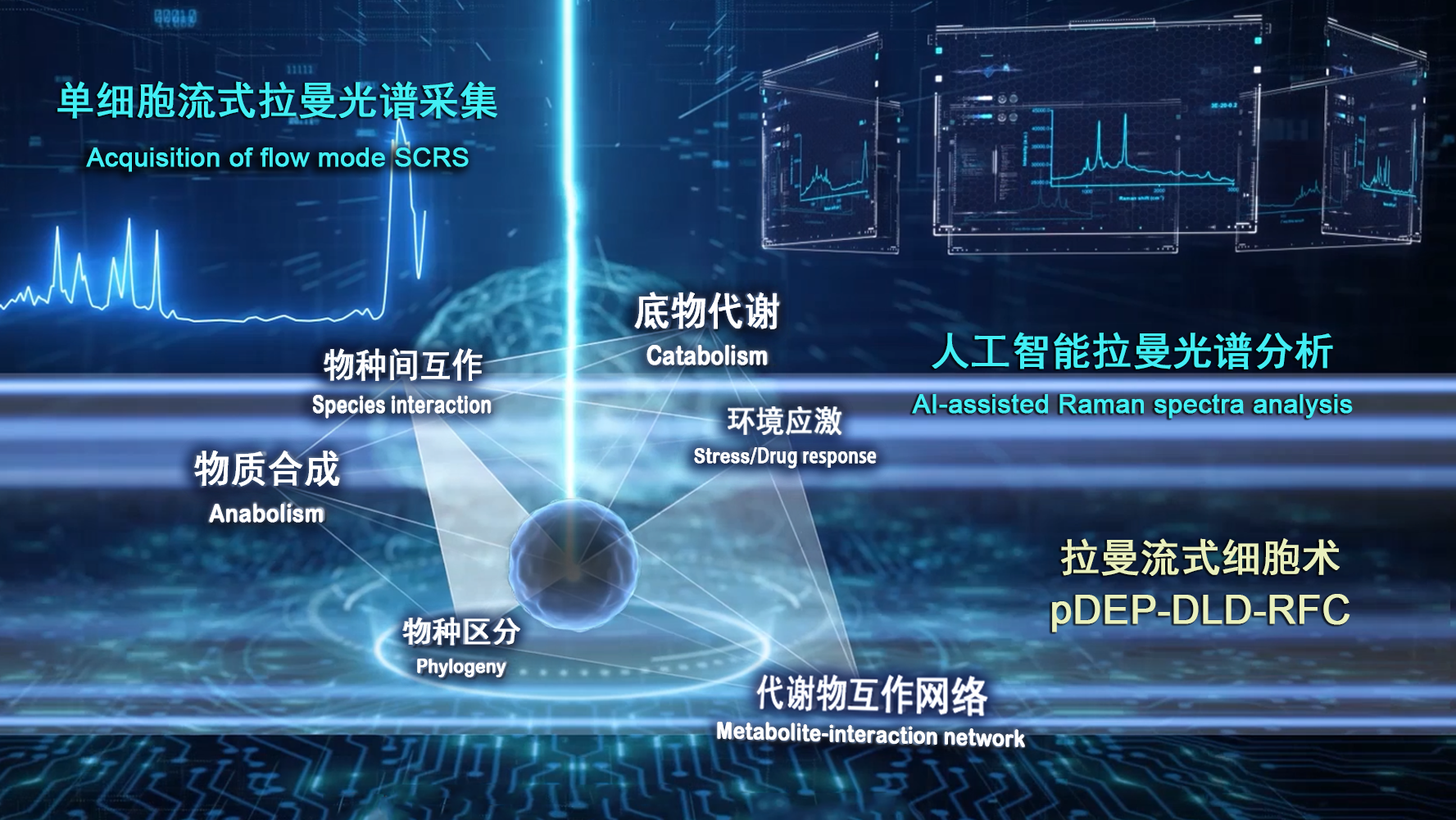
Single-cell metabolic phenome profiling via pDEP-DLD-RFC (Image by LIU Yang)
Analyzing metabolic phenotypes – characteristics that arise from factors like diet, lifestyle, gut microbiome and genetics – involves deep analysis of cellular populations through analytical techniques like mass-spectrometry and fluorescence-based flow cytometry.
These methods, however, involve labelling the cells with fluorescent dyes or destroying them altogether, which has hindered wider deployment. To tackle these challenges, the Chinese research team introduced a new robust flow cytometry platform that does not need to label or destroy the cell.
"The platform's universally applicable, high-throughput nature suggests Raman-based flow cytometry can start to serve a diverse array of novel applications that involve metabolic phenome profiling," said WANG Xixian, study author and associate professor in Single-Cell Center of QIBEBT.
In earlier work, the QIBEBT/CAS research team proposed the "ramanome" concept: a fast, low-cost, high-throughput method for profiling dynamic metabolic features from just one population of genetically identical cells. The approach relies on a collection of single-cell Raman spectra (SCRS), which are structural fingerprints by which molecules can be identified. Each full-spectrum spontaneous SCRS (fs-SCRS) harbors thousands of peaks, which correspond to a specific molecular bond vibration and potentially represent a metabolic phenotype.
A ramanome provides an information-rich metabolic phenome for a given state of a cellular population at single-cell resolution," said Prof. MA Bo from Single-Cell Center of QIBEBT, who led the study. "Ramanomes represent a new non-invasive big-data type for culture-independent, label-free phenomes that can be applied to all cell types."
Despite these promises, acquisition of a ramanome via Raman microscopy is generally a low-throughput method when cells are static, according to the study. For example, collecting a microalgal ramanome can take four hours and only achieve a shallow sampling depth. In contrast, other methods like Raman-based flow cytometry (RFC) and spontaneous Raman flow cytometry are associated with much higher ramanome acquisition throughput. These methods are limited by elevated levels of background noise, poor information content due to narrow spectral range, and low sensitivity due to low spectral resolution.
To combat these limitations, the QIBEBT/CAS research team advanced the ramanome technology by developing and launching a robust Raman-based flow cytometry system for fs-SCRS with high accuracy, high throughput, and stable operation. Called "positive dielectrophoresis-based Raman-activated droplet sorting-induced deterministic lateral displacement-based Raman flow cytometry" (pDEP-DLD-RFC), the new system achieves operating stability with sustained running time for deep sampling of a metabolically heterogeneous cell populations, precise entrapment of fast-moving cells and laser target alignment for efficient spectra acquisition.
pDEP-DLD-RFC consists of a Raman microscope for acquiring SCRS, a microfluidic chip equipped with pumps for controlling buffer flow, a function generator to produce the force that traps and focuses single cells and a relay to control that force. This new technology has been integrated into the instrument called FlowRACS.
Early tests in FlowRACS demonstrated chemical specificity and discrimination accuracy of 99.9% as well as high profiling speeds and throughput.
"For isogenic populations of yeast, microalgae, bacteria like E. coli, and cancer cells, pDEP-DLD-RFC produces deep, highly producible ramanomes that reveal rich, single-cell-resolution phenomes with high throughput and without the need to label the cell with fluorescence probes, thus FlowRACS is a universally applicable tool to profile and sort cells in nature based on their metabolic function," said co-corresponding author Prof. XU Jian, from Single-Cell Center of QIBEBT.
The improvements achieved bypDEP-DLD-RFC promises broad utility in metabolic profiling of cellular systems with applications in medical research and life sciences among other fields.
In future efforts, the research team will explore methods to more efficiently align smaller cells, integrate pDEP-DLD with surface-enhanced Raman scattering to reduce the acquisition time, incorporate a line-focusing strategy to detect multiple cells for SCRS in parallel and streamline single-cell sequencing or cultivation.