Researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) have proposed a new technology, called optical tweezer-assisted pool-screening and single-cell isolation (OPSI) system, which achieves 99.7% purity of sorting target cells, with all done in real-time.
The study was published in Lab on a Chip on Nov. 29.
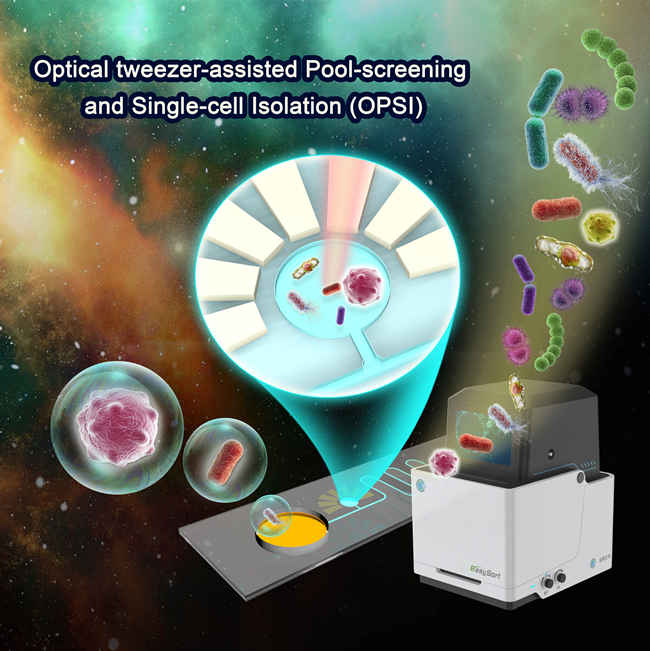
Versatile, facile and low-cost single-cell isolation, culture and sequencing by optical tweezer-assisted pool-screening (Image by LIU Yang)
Current cell-sorting methods cannot effectively sort cells of various sizes while maintaining their viability for future testing. Compared with the currently used methods, the OPSI technology reduces cost and resources consumed. It also saves time, which is of utmost importance when dealing with abnormal cells or pathogens.
Imagine a regular pair of tweezers: they're used for grabbing small, often unwanted, objects such as a stray hair or splinter. An optical tweezer utilizes this same idea, but instead of a metal object, it is a highly focused laser that can hold, manipulate, and move the desired object, which in this case are the target cells.
Being able to pick or "tweeze" out certain cells comes in handy when dealing with cancerous or other target cells and pathogens that need to be further studied. This optical tweezer is utilized on a cell pool confined in a microfluidics chip, which is usually a glass slide with microchannels molded into the material. Once the target cell is identified (usually through targeted fluorescence, Raman imaging or bright-field microscopy), it can easily be packaged in a microdroplet and exported in a "one-cell one-tube" manner for later amplification and analysis.
"Real-time image-based sorting of target cells in a precisely indexed manner is desirable for sequencing or cultivating individual human or microbial cells directly from clinical or environmental samples, however, versatility of existing methods is limited as they are usuallynot broadly applicable to all cell sizes," said XU Teng, paper-first author from Single-Cell Center of QIBEBT.
An artificial test mixture of green-fluorescent protein (GFP) E. coli, non-GFP E. coli and yeast were loaded onto the chip in a 1:1:1 ratio, and quickly the GFP bacteria and yeast were separated.
To further test the efficacy of this method, a mixture using only 0.1% of the GFP E. coli was used, and the fluorescent cells were easily detected and isolated amidst a mixture of other cells of varying sizes.
"The precise isolation and broad spectrum of cell sizes that can be manipulated using OPSI not only allows for easy target cell acquisition but can also greatly reduce the volume required to study the sample," said co-first author LI Yuandong, an engineer at Single-Cell Center of QIBEBT. Isolating and capturing the target cells in microdroplets also maintains a high quality of the cell's information, allowing for more genes to be detected while minimizing the resources needed. "This is of particular importance when it comes to rare or small samples which can easily be consumed entirely in one test which may not even maintain the quality of the sample."
"Taking advantage of the wide-field imaging rather than detecting single cells one by one in flowing stream, the recognition of target cell can be very fast," said co-corresponding author Prof. XU Jian, from Single-Cell Center of QIBEBT. "OPSI also achieves >99.7% target-cell sorting purity and 10-fold elevated speed of 10~20 cells/min."
In the next step, introduction of artificial intelligence-based automatic recognition to this OPSI chip method, along with automatic manipulation steps, may further increase the throughput and greatly broaden the usage of this technology, according to Prof. MA Bo from Single-Cell Center of QIBEBT, who led the study.