When treating acute infections in patients, health care providers must be able to quickly and accurately identify the best antibiotics for fighting the infection.
A research team led by researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) and Peking Union Medical College Hospital has developed an automated system that provides swift, accurate results for determining the best antibiotics at the right dose.
This automated system, the Clinical Antimicrobials Susceptibility Test Ramanometry (CAST-R), is based on Raman microspectroscopy, a spectroscopic technique that can identify specific molecules. It can offer great potential for the treatment of bloodstream infections in humans.
Their findings were published on April 18 in the journal mLife.
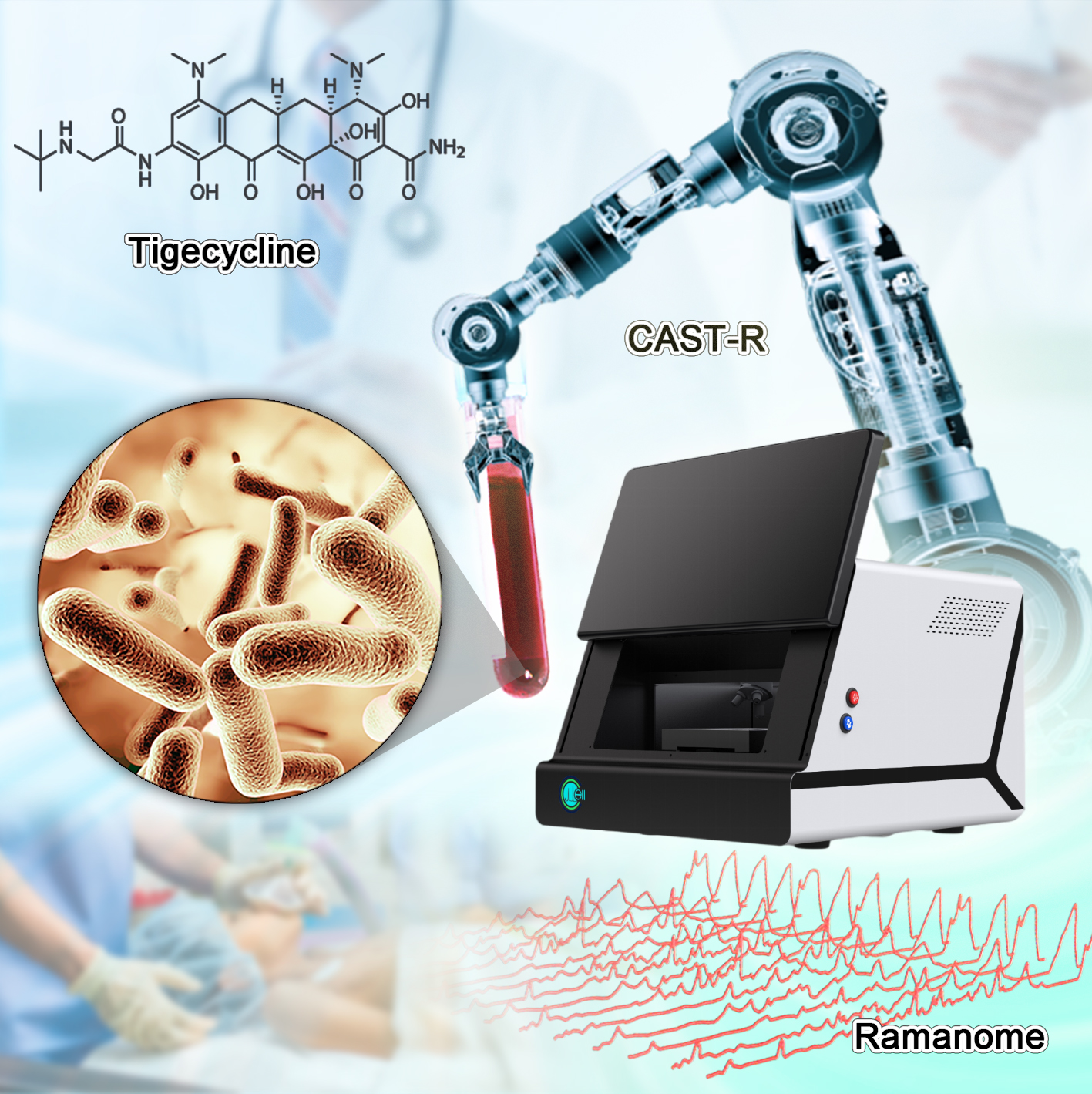
The automated CAST-R workflow supports rapid and accurate treatment of pathogen infections in clinics.
(Image by LIU Yang)
Conventional antimicrobial susceptibility tests (ASTs) have been the essential tool for specifically identifying which antibiotic is most effective in treating a particular bacteria or fungus. But they are time-consuming, labor-intensive and unstable.
Sepsis, a kind of acute blood infections, carries a 20~40% mortality rate, and each hour's delay in obtaining AST report would raise mortality rate by 7.6%. Tigecycline is a last-resort antimicrobial used to treat blood infections. "Unfortunately, resistance to tigecycline has emerged in many blood pathogens," said YANG Qiwen, senior author of the study and Associate Director of Department of Clinical Laboratory, Peking Union Medical College Hospital.
Conventional ASTs for tigecycline have been slow, tedious and even plagued by inaccuracy. The AST of tigecycline has a turn-round time of 36 to 48 hours, from positive blood culture to AST results. "Rapid and reliable methods for tigecycline resistance are urgent, "said ZHU Pengfei, a scientist with QIBEBT, CAS.
So, the research team employed tigecycline as its main model and established an automated system of CAST-R, using the D2O-probed Raman microspectroscopy technology to perform AST for bloodstream infections. CAST-R can help clinicians quickly determine which antibiotic drug to use and how much to prescribe.
The team's process uses a liquid robot for sample pretreatment and a machine learning-based control scheme for acquiring the data and maintaining quality control. Each Single-Cell Raman Spectrum (SCRS) that their process produced contained thousands of Raman peaks providing rich information about cells, kind of like a molecular fingerprint of each cell.
In their tests, the three-hour CAST-R process achieved excellent accuracy and proved to be over ten times faster than the conventional AST process. Their process handled 96 paralleled antibiotic-exposure reactions and produced Raman spectra with quality equivalent to that achieved through the more time-consuming manual process. "The automation, speed, reliability, and broad applicability suggest CAST-R as a clinically valuable AST for bloodstream infections," said REN Lihui, an associate professor at the QIBEBT, CAS.
"The strengths of a SCRS, which also include information richness, single-cell resolution, and ability to couple with downstream single-cell sequencing, have yet to be fully exploited in this study, and that will be our priority ahead," said XU Jian, another senior author of the study and Director of Single-Cell Center at QIBEBT.
The team plans to explore these strengths and further improve the CAST-R process. "As we refine the CAST-R workflow process, our goal is to provide a series of new applications in clinical diagnosis based on microbial single-cell technologies, in order to combat superbugs and support personalized treatment of infections," said YANG Qiwen.